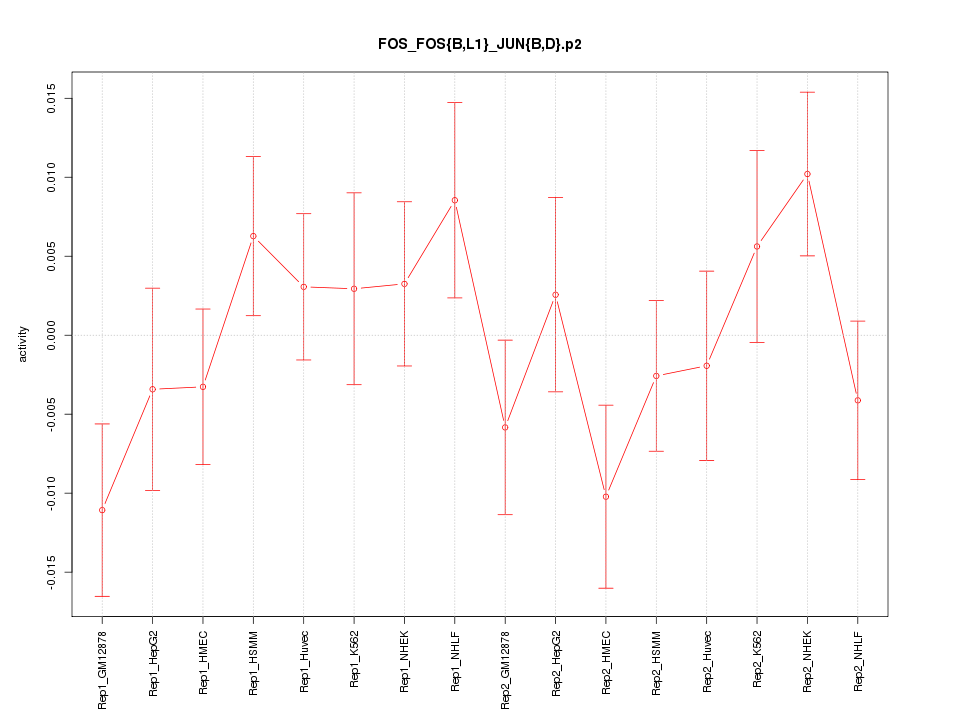
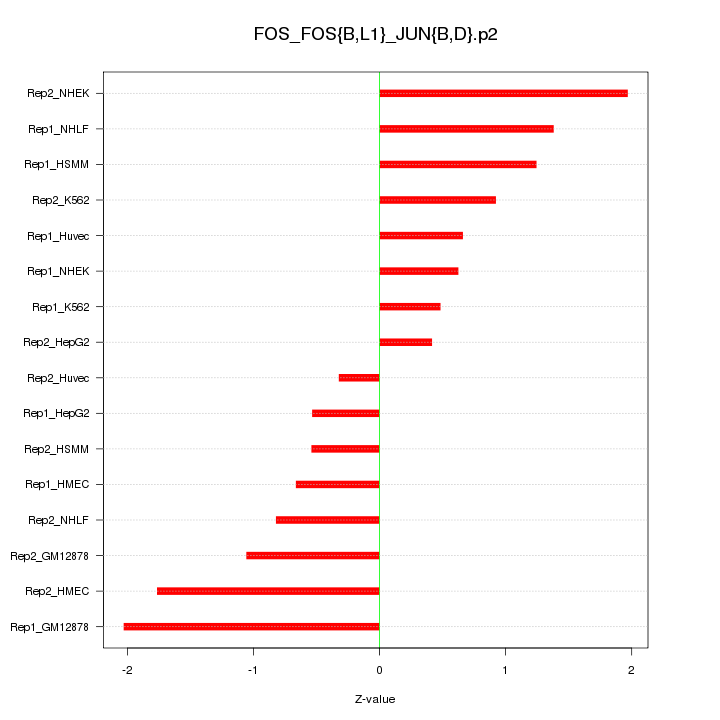
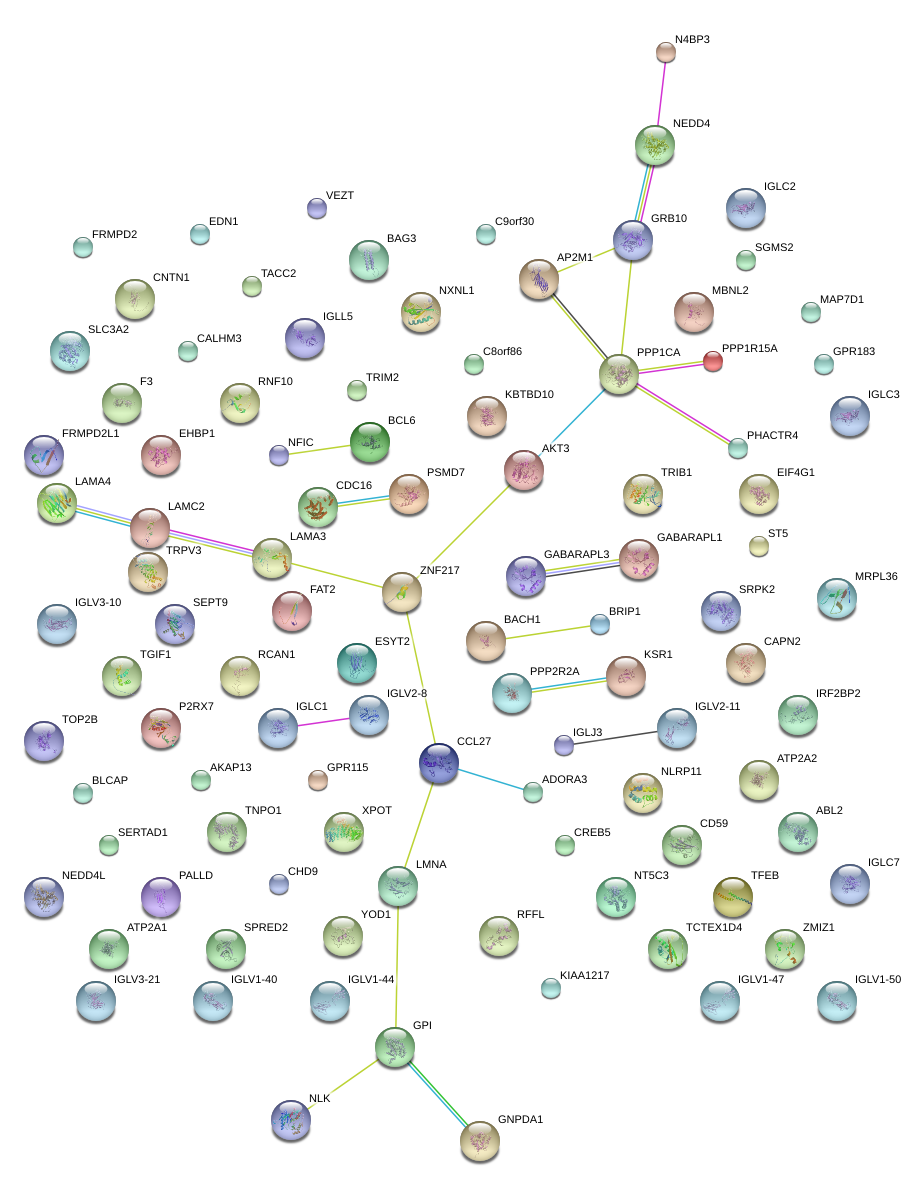
|
chr22_+_21495273
|
2.250
|
|
|
|
|
chr1_-_177378801
|
1.842
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr22_+_21370394
|
1.777
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr10_+_80498782
|
1.720
|
NM_020338
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr22_+_21464980
|
1.575
|
|
|
|
|
chr2_-_65447267
|
1.544
|
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr2_+_87536174
|
1.436
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr2_+_87536106
|
1.397
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr22_+_21407066
|
1.374
|
|
|
|
|
chr2_-_65447370
|
1.280
|
NM_001128210
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr17_+_72958330
|
1.240
|
|
SEPT9
|
septin 9
|
|
chr22_+_21393107
|
1.203
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr1_+_181421984
|
1.193
|
|
LAMC2
|
laminin, gamma 2
|
|
chr17_+_72958207
|
1.187
|
NM_001113496
|
SEPT9
|
septin 9
|
|
chr7_-_104696678
|
1.167
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr1_+_15128882
|
1.139
|
NM_001018001
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr17_+_22823151
|
1.120
|
NM_014238
|
KSR1
|
kinase suppressor of ras 1
|
|
chr1_+_181421796
|
1.065
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr1_+_154351082
|
1.054
|
NM_005572
NM_170707
NM_170708
|
LMNA
|
lamin A/C
|
|
chr22_+_21053972
|
1.027
|
|
|
|
|
chr22_+_21116283
|
0.989
|
|
|
|
|
chr12_+_10256682
|
0.988
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr20_-_51633042
|
0.968
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr6_-_169392771
|
0.924
|
|
|
|
|
chr6_+_47774247
|
0.908
|
NM_153838
|
GPR115
|
G protein-coupled receptor 115
|
|
chr17_+_23393744
|
0.887
|
NM_016231
|
NLK
|
nemo-like kinase
|
|
chr8_-_38505331
|
0.881
|
NM_207412
|
C8orf86
|
chromosome 8 open reading frame 86
|
|
chr1_-_45045543
|
0.848
|
NM_001013632
|
TCTEX1D4
|
Tctex1 domain containing 4
|
|
chr22_+_21552897
|
0.842
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr1_+_36394338
|
0.834
|
NM_018067
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr9_+_102231616
|
0.822
|
NM_001198806
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr19_+_54068373
|
0.803
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr18_+_54039779
|
0.791
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr7_-_50740491
|
0.788
|
NM_001001550
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr1_+_154351162
|
0.784
|
|
LMNA
|
lamin A/C
|
|
chr12_+_94200306
|
0.784
|
|
VEZT
|
vezatin, adherens junctions transmembrane protein
|
|
chr4_+_154397975
|
0.780
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr22_+_20899155
|
0.769
|
|
|
|
|
chr15_-_53996597
|
0.761
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr17_+_23394062
|
0.753
|
|
NLK
|
nemo-like kinase
|
|
chr18_+_54039818
|
0.750
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr19_+_54068300
|
0.741
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr18_+_54039730
|
0.738
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_-_94779727
|
0.731
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr7_+_28692122
|
0.728
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr20_-_35586366
|
0.728
|
NM_001167823
|
BLCAP
|
bladder cancer associated protein
|
|
chr1_+_154351200
|
0.727
|
|
LMNA
|
lamin A/C
|
|
chr13_+_96726415
|
0.716
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr11_-_8695974
|
0.712
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr18_+_3443771
|
0.708
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr22_+_21079355
|
0.695
|
|
|
|
|
chr1_+_181422016
|
0.694
|
|
LAMC2
|
laminin, gamma 2
|
|
chr13_+_96726197
|
0.688
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr10_+_24795446
|
0.682
|
|
KIAA1217
|
KIAA1217
|
|
chr13_-_98757634
|
0.673
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr10_+_24795465
|
0.672
|
|
KIAA1217
|
KIAA1217
|
|
chr7_-_33047042
|
0.672
|
NM_001166118
NM_016489
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr18_+_19706979
|
0.662
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr1_-_205290921
|
0.661
|
NM_018566
|
YOD1
|
YOD1 OTU deubiquinating enzyme 1 homolog (S. cerevisiae)
|
|
chr22_+_21340757
|
0.659
|
|
|
|
|
chr1_-_36589744
|
0.654
|
|
|
|
|
chr5_+_72179685
|
0.643
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr17_-_30414789
|
0.632
|
NM_001017368
|
RFFL
|
ring finger and FYVE-like domain containing 1
|
|
chr22_+_21042091
|
0.617
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr17_+_55269974
|
0.608
|
|
|
|
|
chr2_-_55053398
|
0.606
|
|
|
|
|
chr16_+_51690564
|
0.588
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr11_+_64950402
|
0.581
|
|
|
|
|
chr5_+_72179749
|
0.581
|
|
TNPO1
|
transportin 1
|
|
chr8_+_126511739
|
0.578
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr10_-_49152918
|
0.568
|
NM_001018071
|
FRMPD2
|
FERM and PDZ domain containing 2
|
|
chr1_+_28637247
|
0.563
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr5_+_177473071
|
0.552
|
NM_015111
|
N4BP3
|
NEDD4 binding protein 3
|
|
chr1_+_36394044
|
0.550
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr15_+_84021271
|
0.541
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr4_+_108965167
|
0.541
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr9_-_34652629
|
0.539
|
NM_006664
|
CCL27
|
chemokine (C-C motif) ligand 27
|
|
chr3_-_188936941
|
0.532
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr21_-_34820916
|
0.530
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr2_+_62787534
|
0.516
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr21_+_29593098
|
0.515
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr4_+_169989680
|
0.515
|
NM_001166110
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr10_+_121400926
|
0.511
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr8_+_26206648
|
0.509
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr19_-_61039939
|
0.508
|
NM_145007
|
NLRP11
|
NLR family, pyrin domain containing 11
|
|
chr1_-_111908085
|
0.503
|
NM_001081976
|
ADORA3
|
adenosine A3 receptor
|
|
chr19_-_45623720
|
0.500
|
NM_013376
|
SERTAD1
|
SERTA domain containing 1
|
|
chr3_-_25658829
|
0.495
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr6_+_12398514
|
0.493
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr16_+_72888161
|
0.489
|
NM_002811
|
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7
|
|
chr12_+_119456538
|
0.487
|
|
RNF10
|
ring finger protein 10
|
|
chr19_+_3317575
|
0.485
|
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr1_-_232810111
|
0.481
|
|
IRF2BP2
|
interferon regulatory factor 2 binding protein 2
|
|
chr10_-_105228986
|
0.480
|
NM_001129742
|
CALHM3
|
calcium homeostasis modulator 3
|
|
chr10_+_123862430
|
0.479
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr2_+_170074457
|
0.473
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr12_+_63084317
|
0.471
|
NM_007235
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr3_+_185520744
|
0.469
|
NM_004953
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr16_+_72888231
|
0.466
|
|
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7
|
|
chr21_+_29593088
|
0.466
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr3_+_185521139
|
0.464
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr1_+_221966849
|
0.463
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr11_+_62405736
|
0.462
|
|
SLC3A2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr10_+_121400881
|
0.459
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr19_+_3317589
|
0.459
|
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr17_-_3407780
|
0.457
|
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr1_+_221966835
|
0.454
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr3_+_185377341
|
0.454
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr1_-_242073094
|
0.452
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr12_+_109203412
|
0.452
|
NM_001135765
NM_001681
NM_170665
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr19_-_17432724
|
0.451
|
NM_138454
|
NXNL1
|
nucleoredoxin-like 1
|
|
chr7_-_158304062
|
0.450
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr11_-_33701073
|
0.447
|
NM_001127227
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr6_-_41781655
|
0.445
|
|
TFEB
|
transcription factor EB
|
|
chr6_-_116799548
|
0.443
|
|
LOC100287467
|
hypothetical LOC100287467
|
|
chr16_-_45562840
|
0.442
|
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr12_-_52281050
|
0.439
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr17_-_24069299
|
0.439
|
NM_001142624
NM_001142625
NM_001144943
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr10_+_121400741
|
0.435
|
NM_004281
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr19_+_59107802
|
0.434
|
NM_031896
|
CACNG7
|
calcium channel, voltage-dependent, gamma subunit 7
|
|
chr17_+_65582981
|
0.431
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr6_-_35804337
|
0.427
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr16_+_68157370
|
0.426
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr10_+_70745615
|
0.425
|
NM_033496
|
HK1
|
hexokinase 1
|
|
chr6_+_107066422
|
0.421
|
NM_001624
|
AIM1
|
absent in melanoma 1
|
|
chr19_+_45564549
|
0.421
|
|
PLD3
|
phospholipase D family, member 3
|
|
chr4_+_88076418
|
0.420
|
|
|
|
|
chr18_+_59705918
|
0.418
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr11_+_65026382
|
0.417
|
|
|
|
|
chr22_+_20715340
|
0.415
|
|
|
|
|
chr1_+_221966684
|
0.414
|
NM_001748
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr10_+_121401061
|
0.413
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr21_+_29439768
|
0.409
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr19_+_3317536
|
0.409
|
NM_005597
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr1_+_17507276
|
0.407
|
NM_012387
|
PADI4
|
peptidyl arginine deiminase, type IV
|
|
chr9_-_90456824
|
0.406
|
NM_001100111
|
LOC286238
|
hypothetical protein LOC286238
|
|
chr6_+_138230045
|
0.399
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr3_-_39209075
|
0.398
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr10_+_121401032
|
0.397
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr1_+_211291334
|
0.396
|
|
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1
|
|
chr21_+_37714470
|
0.396
|
NM_130438
NM_001396
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr2_-_73170290
|
0.392
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr3_+_192529567
|
0.391
|
NM_174908
NM_178335
|
CCDC50
|
coiled-coil domain containing 50
|
|
chr3_+_185377430
|
0.387
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr11_+_67107852
|
0.386
|
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr6_+_163904564
|
0.386
|
|
QKI
|
quaking homolog, KH domain RNA binding (mouse)
|
|
chr18_+_59295123
|
0.386
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr8_-_13018123
|
0.385
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr19_-_56215084
|
0.379
|
NM_002776
NM_001077500
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr10_-_31328426
|
0.378
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr1_+_221966951
|
0.378
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr21_+_31953803
|
0.375
|
NM_000454
|
SOD1
|
superoxide dismutase 1, soluble
|
|
chr8_-_95289945
|
0.373
|
NM_004063
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine)
|
|
chr5_+_145297490
|
0.373
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr3_+_142569988
|
0.372
|
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr12_+_13240983
|
0.372
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr17_+_55051818
|
0.369
|
NM_004859
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr1_-_45249603
|
0.368
|
NM_024602
|
HECTD3
|
HECT domain containing 3
|
|
chr12_+_109203526
|
0.367
|
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr10_+_121401076
|
0.366
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr3_+_133855979
|
0.365
|
NM_198329
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr6_+_131190237
|
0.363
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr12_+_13240987
|
0.363
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_+_151025376
|
0.362
|
NM_178353
|
LCE1E
|
late cornified envelope 1E
|
|
chr1_+_211291274
|
0.362
|
|
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1
|
|
chr9_+_131974506
|
0.361
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr21_+_29592984
|
0.361
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr3_-_158703821
|
0.360
|
NM_001167911
NM_001167912
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr6_+_41712907
|
0.360
|
|
MDFI
|
MyoD family inhibitor
|
|
chr7_+_150319076
|
0.358
|
NM_000603
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr7_+_30927439
|
0.358
|
NM_001185060
NM_001185061
NM_001185062
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr4_+_183302105
|
0.358
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr17_-_24068759
|
0.358
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr5_+_179180447
|
0.356
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr15_+_49909147
|
0.355
|
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr2_+_30223372
|
0.355
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr10_-_14090459
|
0.354
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr14_+_102870913
|
0.353
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr7_-_128330615
|
0.351
|
|
KCP
|
kielin/chordin-like protein
|
|
chrX_-_76824726
|
0.351
|
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr5_+_179180509
|
0.347
|
|
SQSTM1
|
sequestosome 1
|
|
chr2_+_30223253
|
0.345
|
NM_001127399
NM_001127400
NM_001127401
NM_016061
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr17_-_3408038
|
0.344
|
NM_145068
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr7_-_98983924
|
0.342
|
|
FAM200A
|
family with sequence similarity 200, member A
|
|
chr22_+_20880159
|
0.342
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr10_+_17310475
|
0.342
|
|
VIM
|
vimentin
|
|
chr12_+_106498763
|
0.342
|
NM_001017523
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr10_+_82163568
|
0.336
|
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr3_+_19963865
|
0.334
|
|
RAB5A
|
RAB5A, member RAS oncogene family
|
|
chr3_-_195571748
|
0.330
|
NM_001135057
NM_130830
|
LRRC15
|
leucine rich repeat containing 15
|
|
chr11_-_82460531
|
0.330
|
NM_014488
|
RAB30
|
RAB30, member RAS oncogene family
|
|
chr14_+_19414230
|
0.329
|
NM_001005501
|
OR4K2
|
olfactory receptor, family 4, subfamily K, member 2
|
|
chr5_+_179180517
|
0.329
|
|
SQSTM1
|
sequestosome 1
|
|
chr1_-_156923111
|
0.327
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr6_-_149847718
|
0.326
|
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr1_-_45249554
|
0.326
|
|
HECTD3
|
HECT domain containing 3
|
|
chr10_+_17310263
|
0.325
|
NM_003380
|
VIM
|
vimentin
|
|
chr2_+_27358800
|
0.321
|
NM_032546
NM_187841
|
TRIM54
|
tripartite motif containing 54
|
|
chr2_+_113591940
|
0.321
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr22_+_21094094
|
0.320
|
|
|
|